
Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, Kanehisa M (1999) KEGG: kyoto encyclopedia of genes and genomes. Rho S, You S, Kim Y, Hwang D (2008) From proteomics toward systems biology: integration of different types of proteomics data into network models. Bioinformatics 21:3448–3449Ĭline MS, Smoot M, Cerami E, Kuchinsky A, Landys N, Workman C et al (2007) Integration of biological networks and gene expression data using cytoscape. Maere S, Heymans K, Kuiper M (2005) BiNGO: a cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21:430–438īader GD, Hogue CW (2003) An automated method for finding molecular complexes in large protein interaction networks. Vailaya A, Bluvas P, Kincaid R, Kuchinsky A, Creech M, Adler A (2005) An architecture for biological information extraction and representation. Nucleic Acids Res 32:D258–D261Īshburner M, Ball CA, Blake JA, Butler H, Cherry JM, Corradi J et al (2001) Creating the gene ontology resource: design and implementation. Harris MA, Clark J, Ireland A, Lomax J, Ashburner M, Foulger R et al (2004) The gene ontology (GO) database and informatics resource.
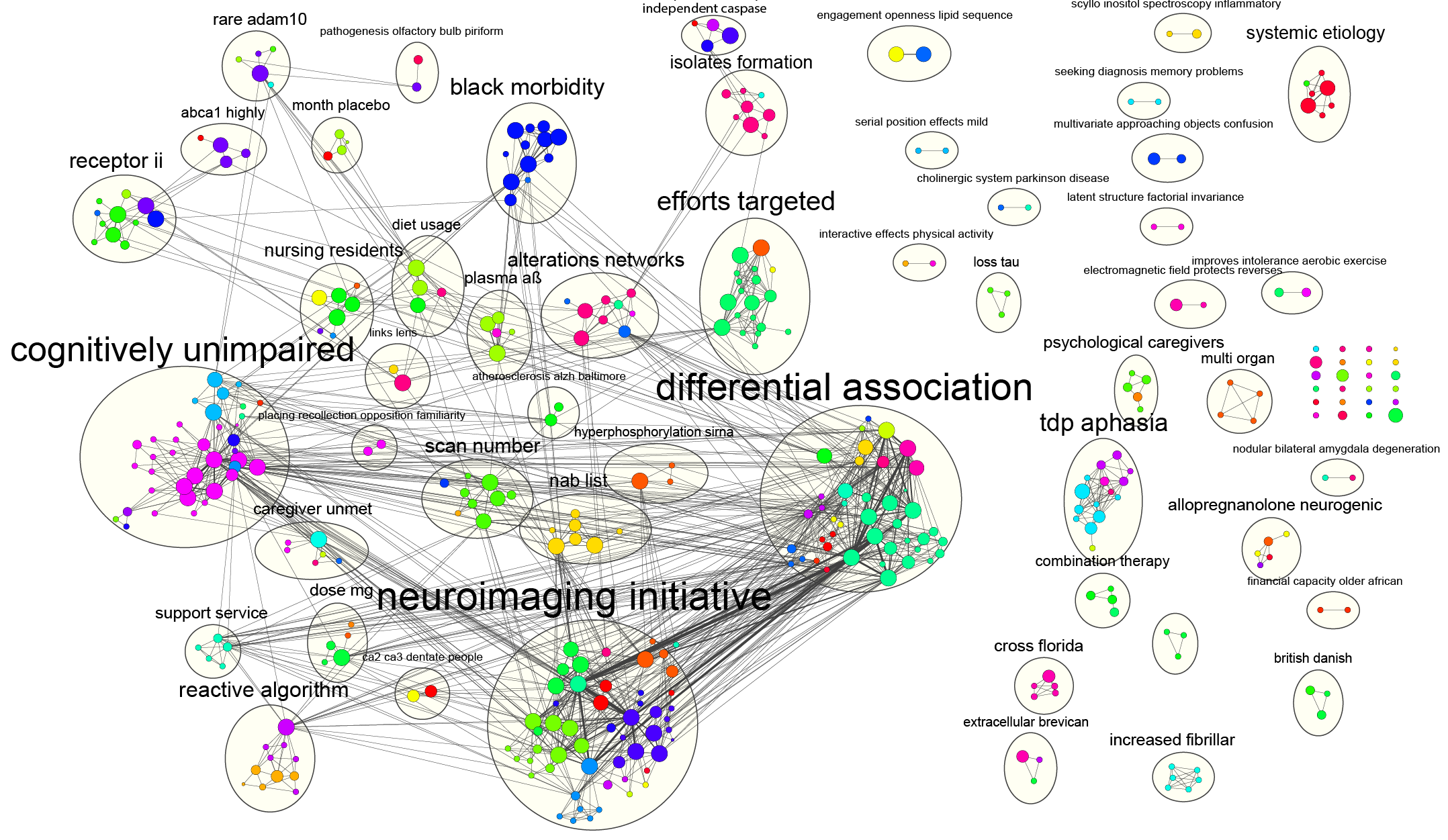
#Cytoscape networks software#
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D et al (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks.
This process is experimental and the keywords may be updated as the learning algorithm improves. These keywords were added by machine and not by the authors. Furthermore, special features of alternative software tools are highlighted in order to assist researchers in the choice of an adequate program for their specific requirements. Additional information about the use of Cytoscape is provided in the Notes section. Utilization of visualization software is exemplified using the widely used Cytoscape tool. Global requirements for such programs are discussed. This chapter focuses on software tools that assist in visual exploration and analysis of biological networks. Obviously, there is a high demand for computer-based assistance for both visualization and analysis of biological data, which are often heterogeneous and retrieved from different sources. Nodes are representations of biological molecules and edges connect the nodes depicting some kind of relationship. In order to represent large biological data sets in an easily interpretable manner, this information is frequently visualized as graphs, i.e., a set of nodes and edges. When you select another group, you can always say: cy.nodes().Substantial progress has been made in the field of “omics” research (e.g., Genomics, Transcriptomics, Proteomics, and Metabolomics), leading to a vast amount of biological data. You now could define a button or something and select the right groups and edges and remove the classes with: cy.nodes("").removeClass(deactivated) Ĭy.edges("").removeClass(deactivate) This way all your nodes are on the graph, they are faded and you have all the edges there. cy.nodes().addClass('deactivated') Ĭy.add('All the edges you have') // Write somthing like species : 1 for first species and so on Additionally you have to give the deactivated property to all nodes. 'font-family': 'Segoe UI,Helvetica Neue,Helvetica,Arial, Verdana',Īfter that you could add all the edges, but you have to add a property to tell which species the edges. You could start with initializin cytoscape with L: // Initialize cytoscape So is that possible to be done in Cytoscape? Thank you in advance. By doing this, one can see how the genes work differently across 3 species. Then you also do the same for zebrafish and cod. When I say, let’s look at human, the network n1 (including both the nodes from l1 and the edges between them) will be hi-lighted (the other genes from the other two species stay there without moving, but faded out as a background). So the background is the all the nodes from L. What I want to visualise is to show 3 networks together. With those 3 networks and 4 lists, I come to Cytoscape. So now I have 3 networks (let’s call them n1, n2, n3), each network for each species. Then I put the 3 gene lists into STRING respectively. Firstly, I search the genes in this list against the genomes of 3 species, to get 3 gene lists for 3 species (let’s call them l1, l2, l3). Of course, the 3 species don’t have all the genes in the list, besides, the interactions between genes are different across 3 species. I have a list of genes (let’s call it L), and I want to see how they work differently in 3 different species (human, zebrafish and cod).


 0 kommentar(er)
0 kommentar(er)
